The use of the zebrafish model system to discover molecular mechanisms of cardiomyopathies
- the causative gene is often unknown
- many variants in novel genes are not functionally validated
- our limited understanding of the molecular mechanisms
- the lack of evidence for specific and effective preventative treatment.
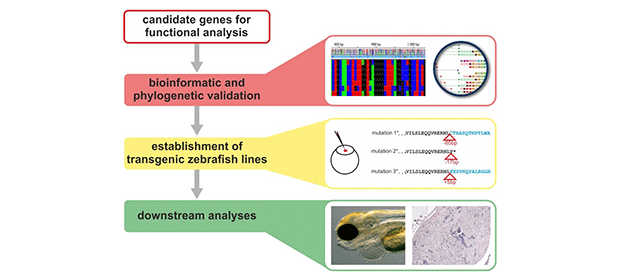
In this project we aim the identification of novel disease-causing genes and the discovery of novel pathways for inherited cardiomyopathies. First, we use whole exome sequencing (WES) in suited families with cardiomyopathies for primary gene discovery. Furthermore, novel candidate gene variants will be selected after bioinformatic analysis and in vitro studies for further disease modelling in zebrafish (Danio rerio) (Fig.1). The project is a collaboration with the Institute of Human Genetics, Würzburg and funded by the IZKF (E-338).
Over the last decade, the zebrafish became an attractive model to study heart development, cardiac function and cardiac effects of genetic variations identified in patients with cardiomyopathies. Due to the high conservation of genes between zebrafish and humans, the zebrafish is well suited to study human cardiovascular disease. Indeed, the direct comparison reveals that at least 71.4% of human protein-coding genes have an orthologue in the zebrafish. In addition to the genetic similarities, zebrafish embryos are fertilized and develop externally, characteristics that permit their manipulation in many ways. Moreover, the use of the Tol2 transposon system permits the generation of novel lines overexpressing mutant genes of interest. Additionally, stable endogenous knock-out or knock-in lines mimicking human genotypes of interest can also be established using the CRISPR/Cas9 technology.
We are in particular interested to characterize mutant lines for cardiac features such as developmental defects, heart failure and sudden death using a broad spectrum of phenotypic assays. The goal is to identify molecular pathways involved in the pathogenesis of cardiomyopathies and potentially select target molecules for future therapeutic interventions. The zebrafish model system also provides the unique perspective to open up the field of high throughput drug/compound discovery by reversal of cardiac fish phenotypes using small molecule drugs.
Figure: Work flow for functional investigations in zebrafish. Following candidate gene identifications, bioinformatic validation are performed to define the correct strategy for each gene individually. Subsequent establishment of transgenic lines are performed either by tol2 overexpression, by CRISPR knock-out or by knock-in methods. Downstream analyses are conducted by a set of different techniques according to the expected heart phenotype.